Basic usage
Usage demonstrations of `SimilaritySearch` with synthetic and real world data
Using the SimilaritySearch package
By Eric S. Téllez
This is a small tutorial showing a minimum example for working with SimilaritySearch it accepts several options that are let to defaults. While this should be enough for many purposes, you are invited to see the rest of the tutorials to take advantage of other features.
using SimilaritySearch
Threads.nthreads()32MatrixDatabase is a required wrapper that tells SimilaritySearch how to access underlying objects since it can support different kinds of objects. In this setup, each column is an object and will be accessed through views using the MatrixDatabase. Since the backend doesn't support appends or pushes, the index can be seen as an static index.
db = MatrixDatabase(randn(2, 10^5))
dist = L2Distance() # squared L2SimilaritySearch.L2Distance()it can use any distance function described in SimilaritySearch and Distances.jl, and in fact any SemiMetric as described in the later package. The index construction is made as follows
G = SearchGraph(; dist, db, verbose=false)
index!(G)SimilaritySearch.SearchGraph{SimilaritySearch.L2Distance, SimilaritySearch.MatrixDatabase{Matrix{Float64}}, SimilaritySearch.AdjacencyLists.AdjacencyList{UInt32}, SimilaritySearch.BeamSearch}
dist: SimilaritySearch.L2Distance SimilaritySearch.L2Distance()
db: SimilaritySearch.MatrixDatabase{Matrix{Float64}}
adj: SimilaritySearch.AdjacencyLists.AdjacencyList{UInt32}
hints: Array{Int32}((116,)) Int32[119, 790, 1126, 1862, 1942, 2109, 2155, 2185, 2278, 2291 … 6510, 6513, 6634, 6650, 6656, 6690, 6692, 6725, 6768, 6782]
search_algo: SimilaritySearch.BeamSearch
len: Base.RefValue{Int64}
verbose: Bool false
this will display a lot of information in the console, since as construction advances the hyperparameters of the index are adjusted. The default optimization takes into account both quality and speed, and tries to adjust to take the best of both. See ParetoRecall in docs.
Once the index is created, the index can solve nearest neighbor queries
Q = MatrixDatabase(randn(2, 100))
k = 30
I, D = searchbatch(G, Q, k)
@show typeof(I) => size(I)
@show typeof(D) => size(D)typeof(I) => size(I) = Matrix{Int32} => (30, 100)
typeof(D) => size(D) = Matrix{Float32} => (30, 100)
I is a matrix of identifiers in db. Each column stores the nearest neighbors (approx.) for the -th colum (object) in Q. The matrix D stores the the corresponding distances for each identifier in I.
Visualizing what we just did
using Plots
@views scatter(db.matrix[1, :], db.matrix[2, :], fmt=:png, size=(600, 600), color=:cyan, ma=0.3, a=0.3, ms=1, msw=0, label="")
for c in eachcol(I)
R = db.matrix[:, c]
@views scatter!(R[1, :], R[2, :], m=:diamond, ma=0.3, a=0.3, color=:auto, ms=2, msw=0, label="")
end
@views scatter!(Q.matrix[1, :], Q.matrix[2, :], color=:black, m=:star, ma=0.5, a=0.5, ms=4, msw=0, label="")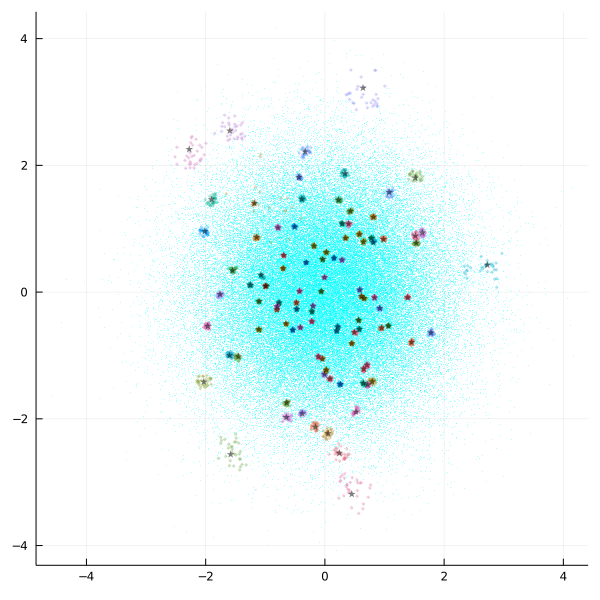
Cyan points identify the dataset while starts are query points. The nearest neighbor points are colored automatically and can repeat, but they come quite close to query points, in dense areas they are even hidding them.
Dependencies
Status `~/Research/SimilaritySearchDemos/site-src/Project.toml`
⌃ [91a5bcdd] Plots v1.15.2
[053f045d] SimilaritySearch v0.10.8
Info Packages marked with ⌃ have new versions available and may be upgradable.